Epigenome Editing Tool Development
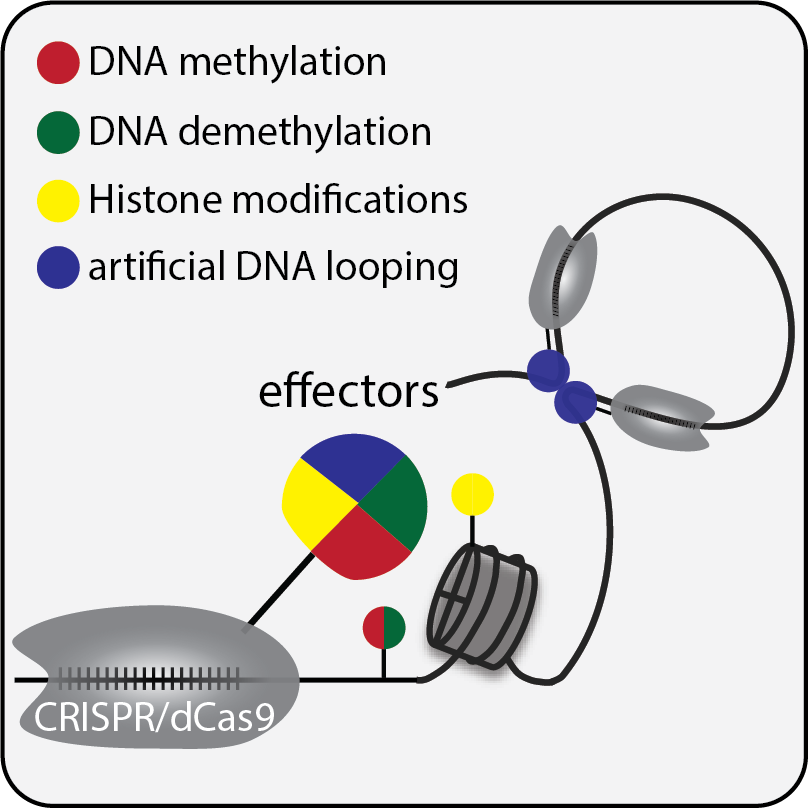
Many breakthroughs in biomedical research were enabled by innovative technology and tool development, which is certainly applicable to the field of epigenetics. Traditionally epigenetics is referred to the study of heritable changes of gene expression without altering the DNA sequence during cell proliferation and development. Several molecular mechanisms including DNA methylation, histone modifications, non-coding RNAs and 3D chromatin structures were identified to be responsible for a variety of intriguing epigenetic phenotypes observed in monocellular organisms such as yeast to multi-cellular organisms like human. For instance, epigenetic modifications undergo dynamic changes upon environmental stimuli, and are considered to enable the genome to interact with the environment (Jaenisch and Bird, 2003). With advances in next generation sequencing technologies, the study of epigenetics has been expanded into epigenome analysis focused on a multitude of chemical modifications and physical properties of the genome at a system level that control the functions of the genome (Bernstein et al., 2007). Numerous epigenetic changes have been associated with biological processes as well as pathological conditions. However, to distinguish the causality and correlation of these changes remains a challenge in this field. Development of epigenome editing tools enables a mechanistic dissection of the functional significance of individual epigenetic events. Our lab pioneered the CRISPR-based DNA methylation editing system including the fusion of a catalytically inactive Cas9 with DNMT3a and TET1 catalytic domains allowing for writing or erasing DNA methylation in the mammalian genome (Liu et al., 2016). The ease of assembly and versatility of CRISPR/dCas9 system to target epigenetic effector protein domains to any given locus allowed this approach to become available to the broad scientific community. We are now expanding the epigenome editing tool box to enable more options for epigenetic manipulations.
References:
Bernstein, B.E., Meissner, A., and Lander, E.S. (2007). The mammalian epigenome. Cell 128, 669-681.
Jaenisch, R., and Bird, A. (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33 Suppl, 245-254.
Liu, X.S., Wu, H., Ji, X., Stelzer, Y., Wu, X., Czauderna, S., Shu, J., Dadon, D., Young, R.A., and Jaenisch, R. (2016). Editing DNA Methylation in the Mammalian Genome. Cell 167, 233-247 e217.