Welcome
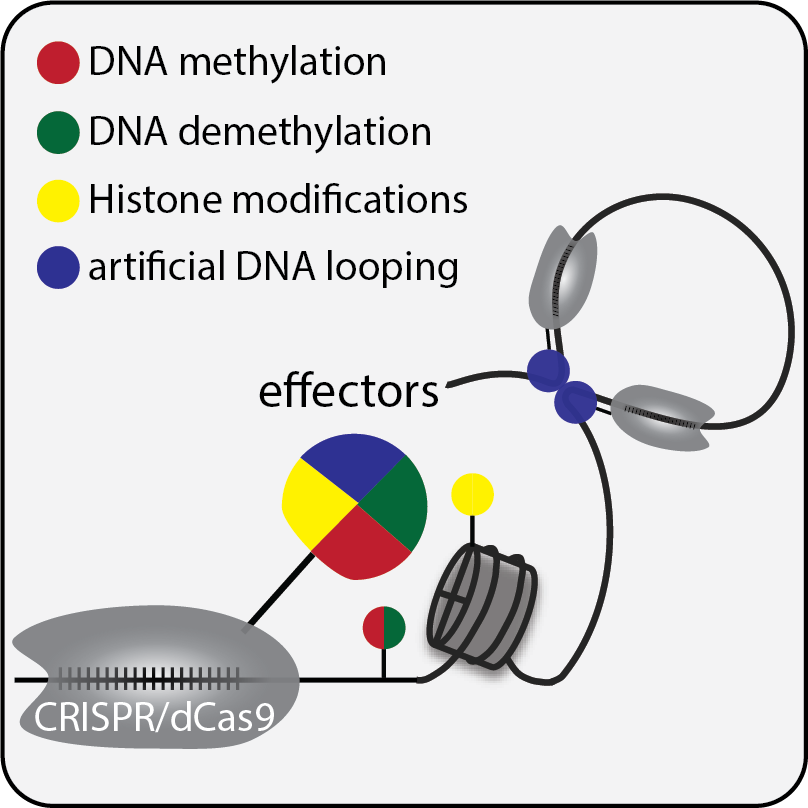
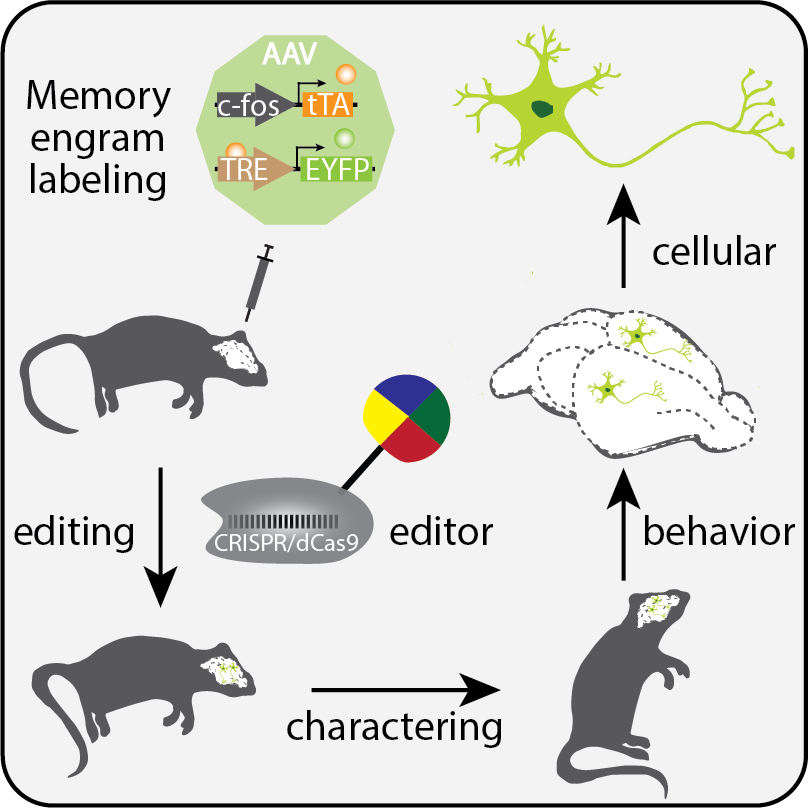
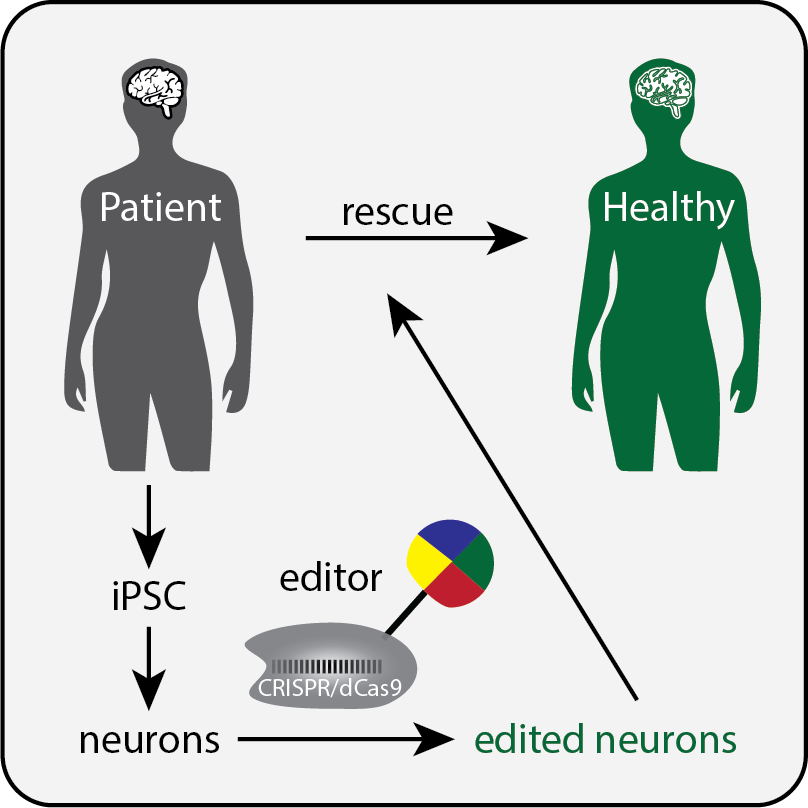
Our laboratory aims to decode the epigenome to understand how genetic information is read in physiology and diseases. We combine molecular tool development with genetic and genomic approaches to explore epigenetic mechanisms underlying normal physiological functions, and to uncover the epigenetic basis of diseases to accelerate the development of therapeutics. We mainly use human ESC- or iPSC-based system and genetically engineered mouse models to tackle these questions.
Members

X. Shawn Liu, PhD Principal Investigator
PyroMark Sequencer Lab Equipment

Maxwell Two Lab Equipment

Xiaonan Guan, PhD Postdoc

Luis Flores, PhD Postdoc

Xinyue (Tracy) Chen Graduate Student

Minglu Wang Undergraduate | Alumni 2020

Alexa Michel Undergraduate | Alumni 2020

Junming Qian Graduate Student

Richard Pan MD-PhD Student

Liana Dawson Undergraduate | Alumni 2021

Bryan Uceda-Alvarez Undergraduate | Alumni 2021-2023

Ji-Hyun Hwang, PhD Postdoc

Rachel Gonzalez, PhD Postdoc

Kitti Rusznák Visiting PhD Student | Alumni 2022

Leslie Yao Undergraduate | Alumni 2022-2023

Jingwei (Jannie) Ren DO Medical Student | Alumni 2021-2023

Kehan Yi Research Associate

Jianxiang (Tom) Ye Rotation Student 2022

Alan Garcia-Epelboim Medical Student

Aadesh Chordia Undergraduate | Alumni 2023

Fengwei Zhan Undergraduate | Alumni 2023

Grace Petrusek Undergraduate

Fanwei Ruan Undergraduate

Isabel McFarland Undergraduate
Publications
Publications since independent in 2020
- Junming Qian and X. Shawn Liu. (2024). CRISPR/dCas9-Tet1 Mediated DNA Methylation Editing. Bio-Protocol 14(8): e4976. PDF
- Liu, Y., Flamier, A., Bell, G.W., Diao, A.J., Whitfield, T., Wang, H., Wu, Y., Schulte, F., Friesen, M., Guo, R., Mitalipova, M., Liu, X.S., Vos, S.M., Young, R.A., Jaenisch, R. (2024). MECP2 directly interacts with RNA polymerase II to modulate transcription in human neurons. Neuron (Accepted for Publication)
- Pedro J. del Rivero Morfin, Diego Scala Chavez, Srinidhi Jayaraman, Lin Yang, Stefanie M. Geisler, Audrey L. Kochiss, Petronel Tuluc, Henry M. Colecraft, Steven O. Marx, X. Shawn Liu, Anjali M. Rajadhyaksha, Manu Ben-Johny (2023). A Genetically Encoded Actuator Boosts L-type Calcium Channel Function in Diverse Physiological Settings. BioRxiv: https://www.biorxiv.org/content/10.1101/2023.09.22.558856v1
- Minglu Wang and X. Shawn Liu. (2023). The emerging field of epigenetic editing: implication for translational purposes for diseases with developmental origin. Perinatal and Developmental Epigenetics Volume 32 in Translational Epigenetics 2023, Pages 355-375, PDF
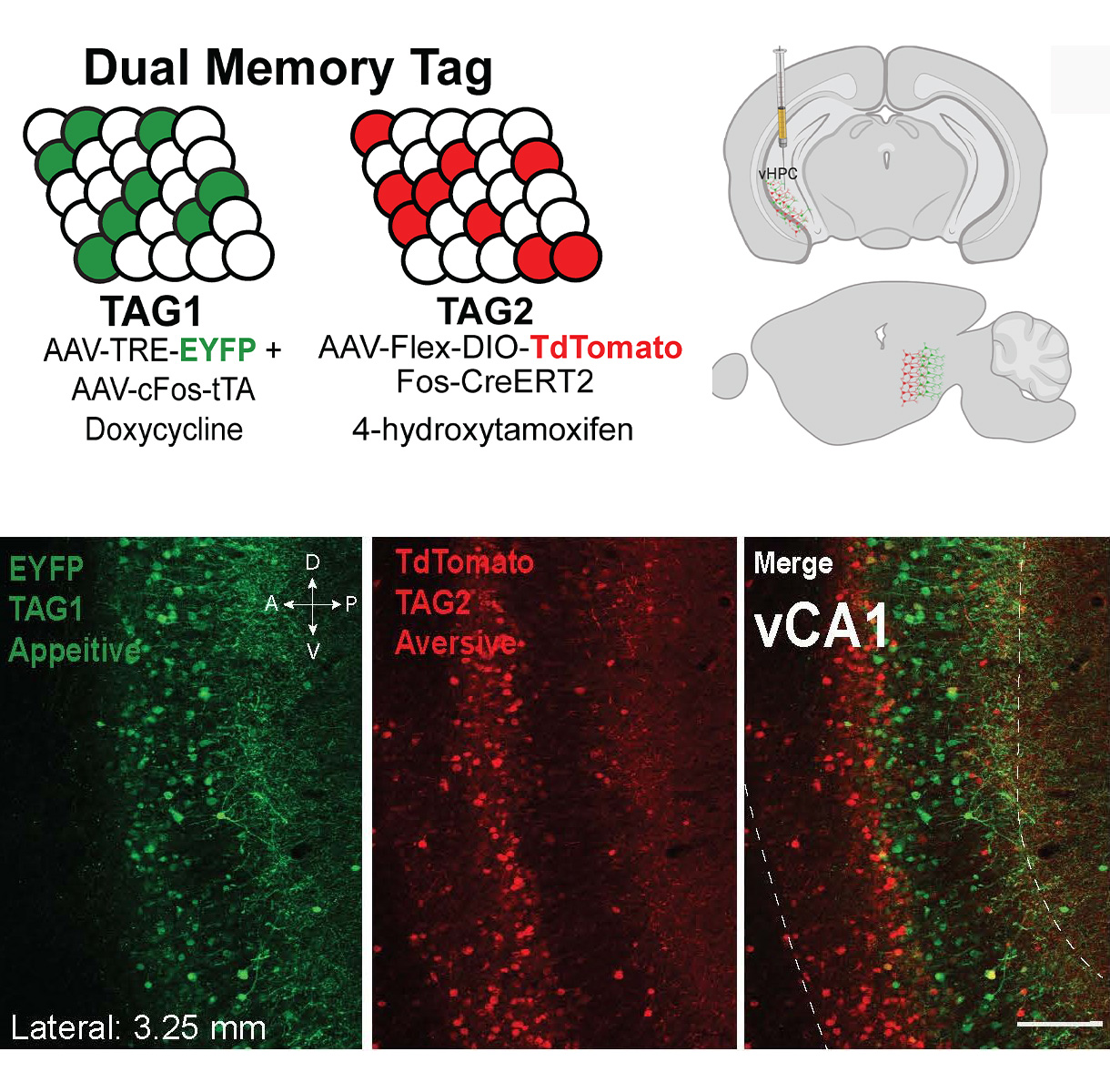
- Jellinger, A.L., Suthard, R.L., Yuan, B., Surets, M., Ruesch, E.A., Caban, A.J., Liu, X.S., Shpokayte, M., Ramirez, S. (2023). Chronic activation of a negative engram induces behavioral and cellular abnormalities. eLife (under review)
- Tomasello, D.L., Barrasa, M.I., Mankus, D., Alarcon, K.I., Lytton-Jean, A.K., Liu, X.S., Jaenisch, R. (2023). Rett Syndrome astrocytes disrupt neuronal activity and cerebral organoid development through transfer of dysfunctional mitochondria. BioRxiv. https://doi.org/10.1101/2023.03.02.530903
-
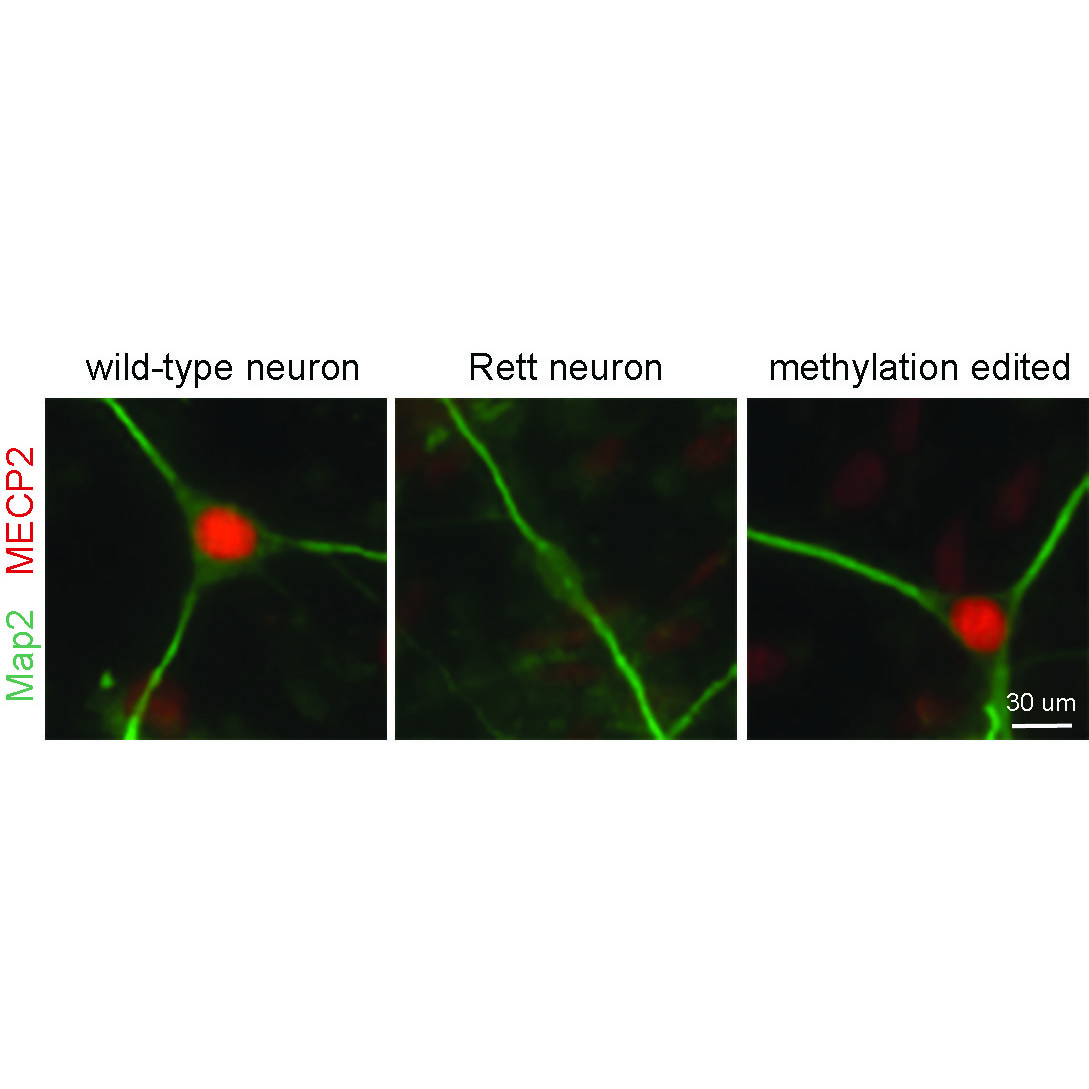
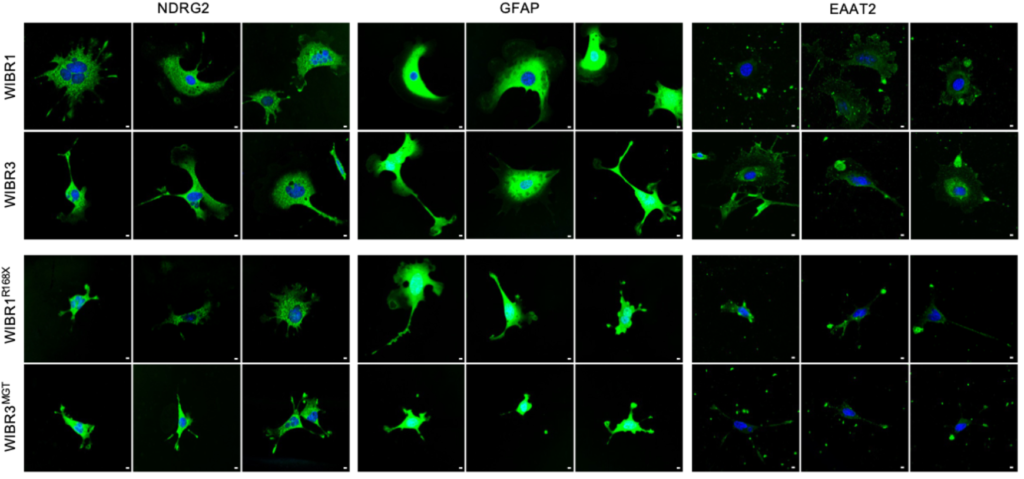
Qian, J., Guan, X., Xie, B., Xu, C., Niu, J., Tang, X., Li, C.H., Colecraft, H.M., Jaenisch, R. Liu, X.S. (2023). Multiplex Epigenome Editing of MECP2 to Rescue Rett Syndrome Neurons. Science Translational Medicine Vol 15, Issue 679. PDF
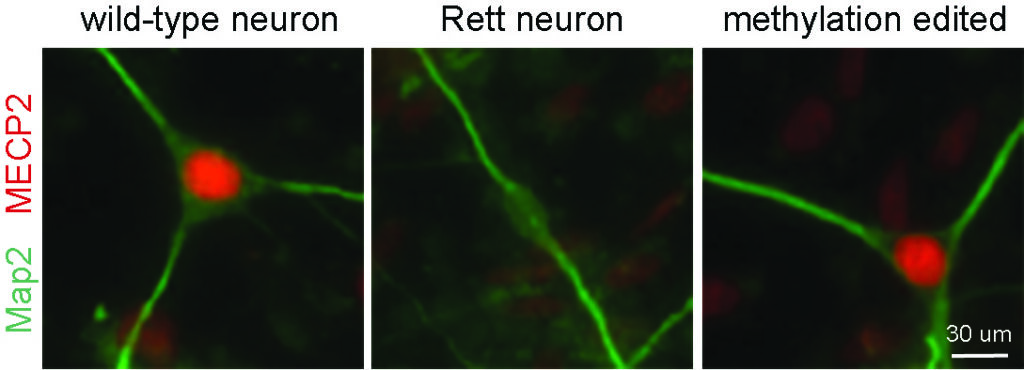
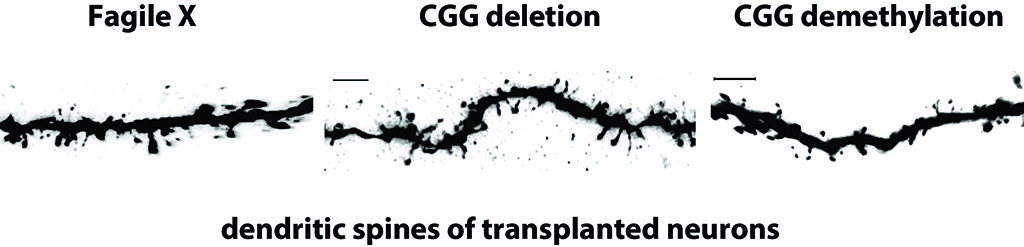
-
Krzisch, M.A., Wu, H., Yuan, B., Whitfield, T.W., Liu, X.S., Fu, D., Garrett-Engele, C.M., Khalil, A.S., Lungjangwa, T., Shih, J., et al. (2022). Fragile X Syndrome Patient-Derived Neurons Developing in the Mouse Brain Show FMR1-Dependent Phenotypes. Biol Psychiatry 93, 71-81. PDF
- Shpokayte, M., McKissick, O., Guan, X., Yuan, B., Rahsepar, B., Fernandez, F., Ruesch, E., Grella, S.L., White, J.A., Liu, X.S.*, Ramirez, S.* (2022). Hippocampal cells segregate positive and negative engrams. (*co-corresponding author). Nature Communication Biology. 5, Article number: 1009. PDF
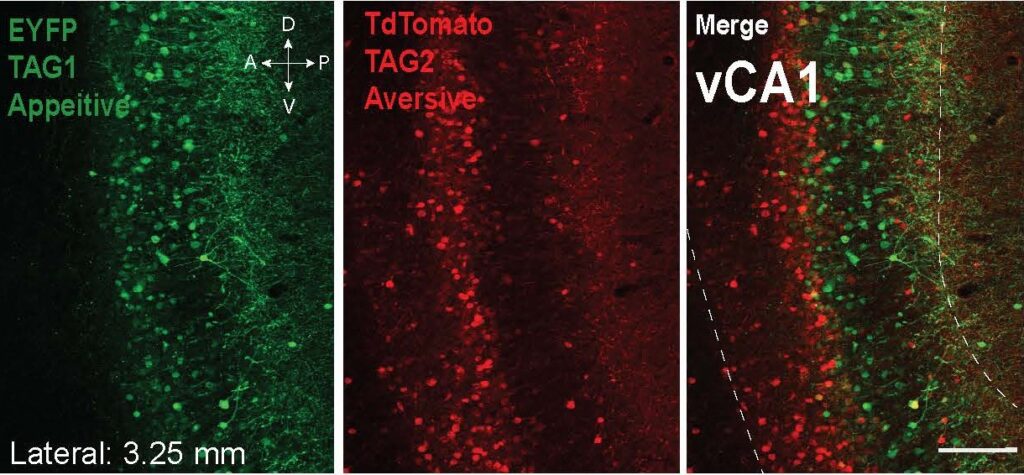
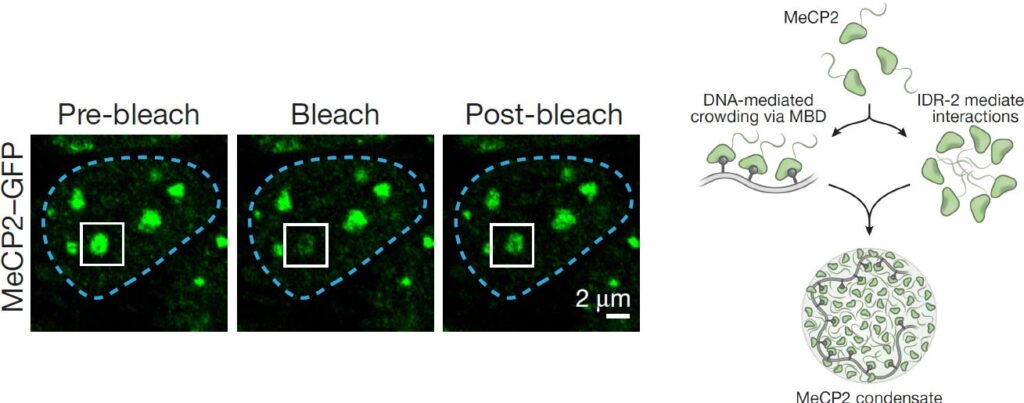
-
Li C.H., Coffey E.L., Dall’Agnese A., Hannett N.M., Tang X., Henninger J.E., Platt J.M., Oksuz O., Zamudio A.V., Afeyan L.K., Schuijers J., Liu X.S., Markoulaki S., Lungjangwa T., LeRoy G., Svoboda D.S., Wogram E., Lee T.I., Jaenisch R., Young, R.A. (2020). MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature 586, 440–444. PDF
Publications before 2020
- Liu, X.S., Jaenisch, R. (2019). Editing the Epigenome to Tackle Brain Disorders. Trends in Neurosciences. (Invited Review) PDF
- Tang, X., Drotar, J., Li, K., Brumm, S.A., Wu, H., Liu, X.S., Wang, J., Gray, N., Sur, M., Jaenisch, R. (2019). Identification of KCC2 Expression Enhancer Compounds as a Basis for Treatment of Rett Syndrome. Science Translational Medicine. 11 (503) PDF
- Liu, X.S., Wu, H., Krzisch, M., Wu, X., Graef, J., Muffat, J., Hnisz, D., Li, C.H., Yuan, B., Vershkov, D., Cacace. A., Young, R.A., and Jaenisch, R. (2018). Rescue of Fragile X syndrome by DNA methylation editing of the FMR1. Cell. 173, 1-14. (Highlighted by Nature Reviews Neuroscience and Nature Reviews Drug Discovery) PDF
- Liu, X.S., Wu, H., Ji, X., Stelzer, Y., Wu, X., Czauderna, S., Shu, J., Dadon, D., Young, R.A., and Jaenisch, R. (2016). Editing DNA methylation in the mammalian genome. Cell. 167, 233-247. (Highlighted by Nature) PDF
- Lee, J.J., Perera, R.M., Wang, H.*, Wu, D.C.*, Liu, X.S.*, Han, S., Fitamant, J., Jones, P.D., Ghanta, K.S., Kawano, S., Nagle, J.M., Deshpande, V., Boucher, Y., Kato, T., Chen, J.K., Willmann, J.K., Bardeesy, N., Beachy, P.A. (2014). Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A. 111, E3091-3100. (*equal contribution) PDF
- Song, B., Liu, X.S., Rice, S., Elzey, B.D., Konieczny, S.F., Ratliff, T., Hazbun, T., Chiorean, E.G., Liu, X., (2012). Plk1 phosphorylation of Orc2 and Hbo1 contributes to gemcitabine resistance in pancreatic cancer. Molecular Cancer Therapeutics. 12, 58-68. PDF
- Liu, W., Wen, Y., Bi, P., Lai, X., Liu, X.S., Liu, X., Kuang, S. (2012). Hypoxia promotes satellite cell self-renewal and enhances the efficiency of myoblast transplantation. Development. 139(16), 2857-65. PDF
- Iliuk, A., Liu, X.S., Xue, L., Liu, X., Tao, W.A. (2012). Chemical Visualization of Phosphoproteomes on Membrane. Mol. Cell Proteomics. 11(9), 629-39. PDF
- Liu, X.S., Song, B., Tang, J., Liu, X. (2012). Plk1 phosphorylates Sgt1 at the kinetochores to promote the timely kinetochore-microtubule attachment. Mol. Cell Biol. 32(19), 4053-67. PDF
- Song B., Liu, X.S., Liu, X. (2012). Polo-like kinase 1 (Plk1): an Unexpected Player in DNA Replication. Cell Division, 7, 3. (invited review) PDF
- Song, B., Liu, X.S., Davis, K., Liu, X. (2011). Plk1 phosphorylation of Orc2 promotes DNA replication under conditions of stress. Mol. Cell Biol., 31(23), 4844-56. PDF
- Song, B., Davis, K., Liu, X.S., Lee, H.G., Smith, M., Liu, X. (2011). Inhibition of Polo-like kinase 1 reduces beta-amyloid-induced neuronal cell death in Alzheimer’s disease. Aging, 9, 846-851. PDF
- Liu, X.S., Song, B., Elzey, B.D., Ratliff, T.L., Konieczny, S.F., Cheng, L., Ahmad, N., Liu, X. (2011). Polo-like Kinase 1 Facilitates Loss of Pten Tumor Suppressor-induced Prostate Cancer Formation. J Biol. Chem., 286, 35795-35800. PDF
- Liu, X.S., Liu, X. (2011). Targeting Plk1 in cutaneous T-cell lymphomas (CTCLs). Cell Cycle. 10(10), 1523. (invited commentary) PDF
- Liu, X.S., Song, B., Liu, X. (2011). The substrates of Plk1, beyond the functions in mitosis. Protein & Cell, 11, 999-1010. (invited review) PDF
- Li, H., Liu, X.S. Yang, X., Song, B., Wang, Y., Liu, X. (2010). Polo-like kinase 1 phosphorylation of p150Glued facilitates nuclear envelope breakdown during prophase. Proc Natl Acad Sci USA, 107, 14633-14638. PDF
- Li, H., Liu, X.S. Yang, X., Wang, Y., Wang, Y., Turner, J.R., Liu, X. (2010). Phosphorylation of CLIP-170 by Plk1 and CK2 promotes timely formation of kinetochore-microtubule attachments. EMBO J, 29, 2953-2965. PDF
- Liu, X.S., Li, H., Song, B., Liu, X. (2010). Polo-like kinase 1 phosphorylation of G2 and S-phase-expressed 1 protein is essential for p53 inactivation during G2 checkpoint recovery. EMBO Rep, 11, 626-632. PDF
- Yang, X., Li, H., Liu, X.S., Deng, A., Liu, X. (2009). Cdc2-mediated phosphorylation of CLIP-170 is essential for its inhibition of centrosome reduplication. J. Biol. Chem., 284, 28775-28782. PDF
Collaborators

Steve Ramirez, PhD Assistant Professor, Boston U

Xuebing Wu, PhD Assistant Professor, Columbia U

Rudolf Jaenisch, MD Professor, Whitehead Institute | MIT

Christine Ann Denny, PhD Associate Professor, Columbia U

Yueqing Peng, PhD Assistant Professor, Columbia U

Neil Shneider, MD Associate Professor, ALS Center | Columbia U

Stavros Lomvardas, PhD Professor, Columbia U